Foundation Model Firsts: What They Enable (and What They Don't)
The past few months have brought a wave of "first foundation models" across scientific domains: spatial proteomics, agriculture, hyperspectral imaging, and solar irradiance forecasting. Each announcement is framed as a breakthrough moment, and in some ways, they are. But the real story isn't about achieving perfection on day one. It's about how these models fundamentally expand the design space for what you can build, test, and deploy next. A foundation model "first" isn't an endpoint—it's an inflection point that raises the baseline and changes what's possible.
What "Firsts" Actually Represent
When a foundation model arrives in a new domain, it doesn't mean the problem is solved. What it means is that someone has successfully trained a shared representation that captures generalizable patterns across that domain's data. These models provide a new baseline: a starting point that's already encoded domain structure, so you don't have to learn everything from scratch. They accelerate iteration by letting teams focus on the delta—the specific application logic, edge cases, or integration challenges—rather than rebuilding feature extraction for every new project.
How "Firsts" Have Shifted Other Domains
We've seen this pattern before. In computer vision, models like CLIP, DINO, and MAE created shared visual representations that work across thousands of tasks without task-specific training. In digital pathology, CTransPath, UNI, and Virchow provided embeddings that transferred across tissue types, stains, and cancer classification tasks, dramatically reducing the labeled data requirements for new applications. In Earth observation, SatMAE and Prithvi enabled researchers to tackle previously inaccessible problems by providing pretrained backbones that work across sensors, geographies, and downstream tasks. Each "first" didn't solve every challenge in its domain—but it changed the economics of what could be attempted next.
New Scientific Capabilities Enabled by FM Firsts
Foundation models unlock capabilities that were impractical or impossible before. Transfer learning becomes feasible: you can take a model trained on one dataset and fine-tune it for a related problem with far less data. Zero-label exploration allows researchers to analyze new samples using embeddings without needing annotations upfront. Multimodal fusion becomes easier when models learn aligned representations across imaging modalities or sensor types. Standardization emerges as teams converge on shared architectures and feature spaces, making it simpler to reproduce results and compare approaches. Perhaps most importantly, these models reveal domain complexity—exposing where variability exists, where current representations fail, and what the next generation of models needs to address.
Domain Examples: What's Now Possible
In spatial proteomics, foundation models create embeddings that capture tissue architecture, helping researchers identify biomarkers, understand batch effects, and compare samples across experiments without manually engineering spatial features. In agriculture, they enable cross-crop transfer learning and large-scale phenotyping, making it practical to monitor diverse crop types and growth conditions without training separate models for every species. In hyperspectral imaging, they support sensor-agnostic modeling—meaning a single model can generalize across different instruments—and unlock applications in materials science, crop stress detection, and mineral mapping. In space weather forecasting, foundation models create unified embeddings across multiple solar observation instruments—including atmospheric imaging and magnetic field data—enabling improved forecasting of solar wind speed and extreme ultraviolet irradiance that weren't feasible with instrument-specific approaches.
What FM Firsts Don't Do
But let's be clear: these models don't magically fix distribution shift, guarantee ROI, or eliminate the need for domain-specific validation. They don't work perfectly out of the box on every edge case. They don't bypass the hard work of understanding your deployment environment, labeling critical failure modes, or building robust data pipelines. What they do is change the opportunity space—opening up projects that were previously too expensive, too data-hungry, or too technically risky to pursue.
The Real Value of a "First"
A first foundation model isn't perfection. It's an enabling event. It raises the baseline, expands the design space, and makes new experiments tractable. The question isn't whether these models solve everything—it's what they make possible that wasn't before.
Want clarity on where foundation models actually create ROI — and where they don't?
Book a Pixel Clarity Call and I'll help you identify the highest-leverage applications for your data, team, and domain.
- Heather |
|
 |
|
Vision AI that bridges research and reality
— delivering where it matters
|
|
|
|
|
|
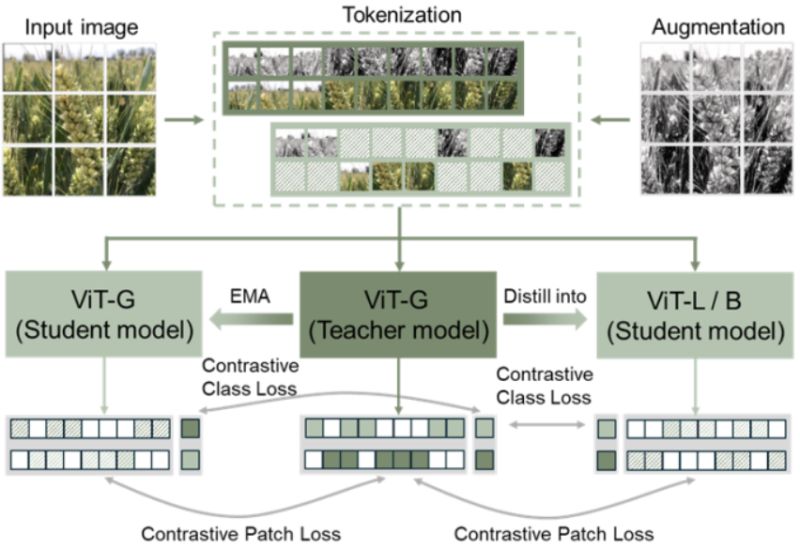 |
Research: Foundation Model for Agriculture
FoMo4Wheat: Toward reliable crop vision foundation models with globally curated data
Foundation models trained on ImageNet work remarkably well for many computer vision tasks. But agriculture presents a unique challenge: fine, variable canopy structures interacting with fluctuating field conditions create a distribution shift that general-domain models struggle to handle.
Bing Han et al. built FoMo4Wheat, one of the first crop-specific vision foundation models, and demonstrated that domain-specific pretraining matters more than we might think.
𝐓𝐡𝐞 𝐝𝐚𝐭𝐚𝐬𝐞𝐭:
ImAg4Wheat is the largest and most diverse wheat image dataset to date: 2.5 million high-resolution images collected over a decade at 30 global sites, spanning more than 2,000 genotypes and 500 environmental conditions. This scale and diversity is critical for capturing the phenotypic variation that agricultural AI systems need to handle.
𝐊𝐞𝐲 𝐟𝐢𝐧𝐝𝐢𝐧𝐠𝐬:
- FoMo4Wheat consistently outperforms state-of-the-art general-domain models (like DINOv2) across 10 in-field vision tasks at both canopy and organ levels
- Achieves strong performance with dramatically less labeled data (e.g., only 30% of training data needed to match SOTA on growth stage and disease classification)
- Despite being trained exclusively on wheat, shows robust cross-crop transfer to rice and accurate differentiation of multiple crop species from weeds
- Demonstrates superior feature representations with clearer clustering of plant organs and disease symptoms
𝐖𝐡𝐲 𝐭𝐡𝐢𝐬 𝐦𝐚𝐭𝐭𝐞𝐫𝐬:
This work validates a path forward for agricultural computer vision: domain-specific foundation models trained on carefully curated, diverse datasets from the target domain. The cross-crop generalization suggests we may be able to build a universal crop foundation model—but getting there requires starting with deep expertise in specific crops first.
Both the models and dataset are open source, charting a collaborative path toward more reliable field-based crop monitoring.
Code and models
Demo |
|
|
|
|
|
 |
Insights: Regulated AI
Why Your Pathology AI May Never Reach Patients
Even the smartest model will stall—if it isn’t built for regulatory pathways.
Many promising models in computational pathology never make it into clinical diagnostics, trials, or drug development pipelines—not because they underperform, but because they’re not built for regulatory scrutiny.
𝐏𝐡𝐚𝐫𝐦𝐚‑𝐫𝐞𝐚𝐝𝐲 𝐀𝐈 𝐢𝐬 𝐚 𝐫𝐞𝐠𝐮𝐥𝐚𝐭𝐞𝐝 𝐩𝐫𝐨𝐝𝐮𝐜𝐭.
To be used in clinical diagnostics, drug development, clinical trials, or as a companion diagnostic, it needs:
- Traceability from code to result
- Clinical validation aligned with Good Clinical Practice and real-world populations
- Risk and failure mode analysis
- Structured documentation (design controls, version history, auditability)
🧪 Example: A team develops a promising subtype classifier. But when a pharma partner requests validation aligned with Good Clinical Practice (GCP) and regulatory reporting standards, the team can’t produce versioned documentation or a risk file. Trust breaks down—and the collaboration dies before the first trial.
Regulatory paths differ: diagnostics need CLIA or FDA clearance, CDx tools require co-development alignment, and trial tools face GCP and IRB oversight.
This isn’t about red tape—it’s about credibility, safety, and the ability to influence high-stakes decisions.
Pharma partners and regulators don’t just want models—they want evidence they can trust.
If you're aiming for trial integration or CDx approval, this is the bar.
𝐒𝐨 𝐰𝐡𝐚𝐭? Without regulatory foresight, even brilliant models die at the translational finish line.
𝐓𝐚𝐤𝐞𝐚𝐰𝐚𝐲: Good performance is just the start. Without regulatory scaffolding, your model may never leave the lab—or help a single patient.
💡 Developer tip: Version everything. Justify key decisions. Build like someone will audit it.
📣 What’s the first regulatory step you wish you’d taken earlier?
Leave a comment |
|
|
|
|
|
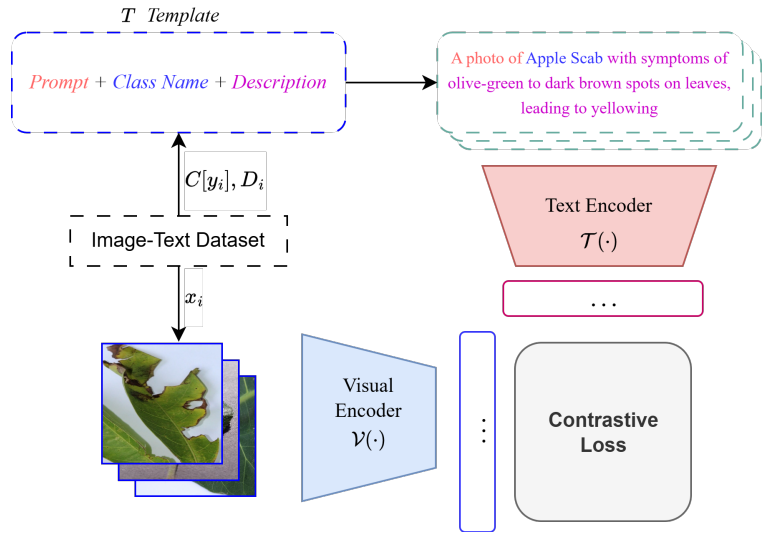 |
Research: VLM for Leaves
A Vision-Language Foundation Model for Leaf Disease Identification
Identifying plant diseases early is critical for food security—but current AI models struggle to generalize across different crops and lack the ability to combine visual and textual information effectively.
Why this matters: Most existing deep learning approaches for plant disease identification focus on single crops and rely solely on images, missing the rich contextual information that text can provide. Plus, they're often pretrained on general datasets like ImageNet, which don't capture agricultural domain knowledge.
Key innovations from Khang Nguyen Quoc et al.:
- SCOLD (Soft-target COntrastive learning for Leaf Disease identification) - Think of it as a softer version of CLIP, specifically designed for agriculture. It's trained on 186k+ agricultural image-caption pairs covering 97 disease concepts across 22 crop species
- Context-Aware Soft Targets - Instead of CLIP's hard one-hot labels that treat all non-matching pairs equally, SCOLD uses soft labels that account for semantic similarity (e.g., diseases affecting the same crop type or sharing symptoms), reducing overconfidence and improving generalization
- Enhanced text descriptions - Disease symptom information enriches simple class labels, creating more nuanced image-text alignment than typical category names
- Strong performance with fewer parameters - SCOLD outperforms OpenAI CLIP, BioCLIP, and SigLIP2 on zero-shot and few-shot classification tasks while maintaining a competitive model size
The model achieved 95%+ accuracy on image-text retrieval and showed robust performance across out-of-distribution crop datasets—even reaching 98%+ on some disease types in few-shot settings.
This represents meaningful progress toward practical AI tools for precision agriculture, especially in scenarios with limited labeled data.
|
|
|
|
|
|
 |
Insights: AI Product Development
What If We Designed Pathology AI for the Real World First?
What if your AI project started with the clinician—not the code?
📍 The real-world pipeline begins in the clinic.
Most pathology AI starts in a lab sandbox—only to struggle when faced with messy, real-world conditions.
But clinical success demands a different approach:
– Anchor development in real clinical pain points—not just in what’s convenient to model
– Design for workflow integration, not just model performance
– Align with regulatory and adoption requirements from the outset
It’s not just about accuracy. It’s about usability, trust, and deployment.
🤝 Co-design matters.
Pathologists, engineers, and product leaders should shape tools together—along with regulatory, QA, and operations teams.
We need to shift incentives—
From novel architectures and benchmark wins → to tools that actually get used.
So what?
Until we build for the clinic from day one, even the smartest models will stay stuck in pilot purgatory.
💬 What would a clinic-first AI pipeline look like in your world—and who should be in the room?
Leave a comment |
|
|
|
|
|
|
Enjoy this newsletter? Here are more things you might find helpful:
Pixel Clarity Call - A free 30-minute conversation to cut through the noise and see where your vision AI project really stands. We’ll pinpoint vulnerabilities, clarify your biggest challenges, and decide if an assessment or diagnostic could save you time, money, and credibility.
Book now |
|
|
|
Did someone forward this email to you, and you want to sign up for more? Subscribe to future emails
This email was sent to _t.e.s.t_@example.com. Want to change to a different address? Update subscription
Want to get off this list? Unsubscribe
My postal address: Pixel Scientia Labs, LLC, PO Box 98412, Raleigh, NC 27624, United States |
|
|
|
|