 |
Research: Vision-Language Model for Whole Slides
CPath-Omni: A Unified Multimodal Foundation Model for Patch and Whole Slide Image Analysis in Computational Pathology
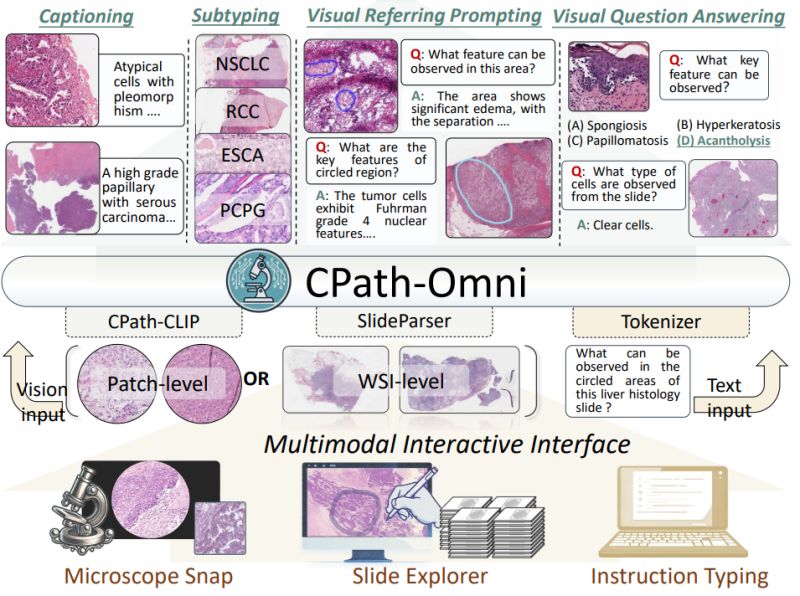
Imagine an AI assistant that can seamlessly analyze everything from individual tissue patches under a microscope to entire gigapixel whole slide images, answering questions like "What key features can be observed in this circled region?" or generating comprehensive pathology reports.
Yuxuan Sun et al. introduced CPath-Omni at CVPR 2025 - the first 15 billion parameter AI model to unify patch-level and whole slide image analysis in computational pathology, bringing us closer to a true "one-for-all" diagnostic assistant.
𝐓𝐡𝐞 𝐟𝐫𝐚𝐠𝐦𝐞𝐧𝐭𝐚𝐭𝐢𝐨𝐧
𝐜𝐡𝐚𝐥𝐥𝐞𝐧𝐠𝐞: Current pathology AI systems are split between patch-level models for detailed tissue analysis and separate whole slide image models for broader diagnostic tasks. This fragmentation leads to redundant development efforts and prevents knowledge transfer between different scales of analysis, limiting the integration of learned patterns across the pathology workflow.
𝐊𝐞𝐲 𝐢𝐧𝐧𝐨𝐯𝐚𝐭𝐢𝐨𝐧𝐬:
∙ 𝐂𝐏𝐚𝐭𝐡-𝐂𝐋𝐈𝐏: Novel pathology CLIP model combining DINOv2-based Virchow2 with traditional CLIP, using a large language model as text encoder for superior alignment
∙ 𝐔𝐧𝐢𝐟𝐢𝐞𝐝
𝐚𝐫𝐜𝐡𝐢𝐭𝐞𝐜𝐭𝐮𝐫𝐞: Single model handling both patch analysis and gigapixel whole slide images up to 100,000 × 100,000 pixels
∙ 𝐅𝐨𝐮𝐫-𝐬𝐭𝐚𝐠𝐞 𝐭𝐫𝐚𝐢𝐧𝐢𝐧𝐠: Progressive learning from patch-based pretraining to mixed patch-WSI training for knowledge transfer
∙ 𝐌𝐮𝐥𝐭𝐢-𝐭𝐚𝐬𝐤 𝐜𝐚𝐩𝐚𝐛𝐢𝐥𝐢𝐭𝐢𝐞𝐬: Classification, visual question answering, captioning, and visual referring prompting across both scales
𝐖𝐡𝐲 𝐭𝐡𝐢𝐬 𝐦𝐚𝐭𝐭𝐞𝐫𝐬: CPath-Omni achieved state-of-the-art performance on 39
out of 42 datasets across seven diverse tasks, even surpassing human pathologist performance on some benchmarks (72.4% vs 71.8% on PathMMU). More importantly, it demonstrates that knowledge learned at the patch level can effectively enhance whole slide image understanding, suggesting a path toward more efficient and comprehensive pathology AI systems.
This unified approach could streamline pathology workflows and make advanced AI diagnostics more accessible in resource-limited clinical settings.
|
|
|
|
|
|
Research: Spatial Transcriptomics
Revolutionizing Drug Discovery: Integrating Spatial Transcriptomics with Advanced Computer Vision Techniques
Understanding where genes are expressed within tissues could be key to finding new drug targets. But extracting actionable insights from this complex, high-dimensional data remains a significant challenge.
Zichao Li, et al. tackle this challenge by combining spatial transcriptomics with computer vision techniques to improve therapeutic target identification in drug discovery.
𝗪𝗵𝗮𝘁 𝗶𝘀 𝗦𝗽𝗮𝘁𝗶𝗮𝗹 𝗧𝗿𝗮𝗻𝘀𝗰𝗿𝗶𝗽𝘁𝗼𝗺𝗶𝗰𝘀?
Spatial transcriptomics maps gene expression within intact tissues, showing not just which genes are
active but exactly where in the tissue they're expressed. This technology bridges genomics and histology, revealing cellular interactions and tissue architecture that traditional methods miss. However, the resulting datasets are extremely high-dimensional and noisy, making analysis computationally challenging.
𝗧𝗵𝗲 𝗔𝗽𝗽𝗿𝗼𝗮𝗰𝗵:
The researchers developed a framework that combines three key components:
• 𝗨-𝗡𝗲𝘁 𝘀𝗲𝗴𝗺𝗲𝗻𝘁𝗮𝘁𝗶𝗼𝗻: Divides gene expression regions into distinct cellular compartments for more precise analysis
• 𝗚𝗿𝗮𝗽𝗵 𝗻𝗲𝘂𝗿𝗮𝗹 𝗻𝗲𝘁𝘄𝗼𝗿𝗸𝘀: Captures spatial relationships between tissue
regions that traditional methods miss
• 𝗠𝘂𝗹𝘁𝗶-𝘁𝗮𝘀𝗸 𝗹𝗲𝗮𝗿𝗻𝗶𝗻𝗴: Simultaneously predicts disease-specific biomarkers and classifies tissue regions
𝗞𝗲𝘆 𝗥𝗲𝘀𝘂𝗹𝘁𝘀:
Testing on mouse brain and human breast cancer datasets showed substantial improvements:
• 92.3% accuracy vs 85.7% for standard U-Net approaches
• Better performance in noisy conditions (85.2% accuracy at 20% noise vs 71.3% for baselines)
• Superior identification of disease-specific biomarkers across multiple metrics
𝗜𝗺𝗽𝗹𝗶𝗰𝗮𝘁𝗶𝗼𝗻𝘀 𝗳𝗼𝗿 𝗗𝗿𝘂𝗴
𝗗𝗲𝘃𝗲𝗹𝗼𝗽𝗺𝗲𝗻𝘁:
Traditional drug discovery methods are time-consuming and often lack precision. By enabling more accurate identification of therapeutic targets from spatial gene expression data, this approach could accelerate the discovery pipeline. The integration of spatial context is particularly valuable for understanding complex diseases like cancer, where cellular interactions within the tumor microenvironment are critical.
This work demonstrates how computer vision techniques originally developed for image analysis can be adapted to unlock insights from biological data, potentially leading to more targeted and effective therapies.
|
|
|
|
|
|
Enjoy this newsletter? Here are more things you might find helpful:
Modeling Roadmap -- Are you afraid your computer vision project will turn into an endless stream of failed experiments? What if you had a clear strategy for implementation? My Modeling Roadmap is a strategic plan outlining the components you’ll need to make your computer vision project a success using best practices for your type of imagery and bringing in cutting-edge research where needed.
Apply now
|
|
|
Did someone forward this email to you, and you want to sign up for more? Subscribe to future emails
This email was sent to _t.e.s.t_@example.com. Want to change to a different address? Update subscription
Want to get off this list? Unsubscribe
My postal address: Pixel Scientia Labs, LLC, PO Box 98412, Raleigh, NC 27624, United States
|
|
|
|