 |
Insights: Foundation Models
Why Do Pathology and Earth Observation Foundation Models Look So Different?
𝘞𝘩𝘺 𝘥𝘰 𝘗𝘢𝘵𝘩𝘰𝘭𝘰𝘨𝘺 𝘢𝘯𝘥 𝘌𝘖 𝘧𝘰𝘶𝘯𝘥𝘢𝘵𝘪𝘰𝘯 𝘮𝘰𝘥𝘦𝘭𝘴 𝘭𝘰𝘰𝘬 𝘭𝘪𝘬𝘦 𝘵𝘩𝘦𝘺'𝘳𝘦 𝘧𝘳𝘰𝘮 𝘥𝘪𝘧𝘧𝘦𝘳𝘦𝘯𝘵 𝘸𝘰𝘳𝘭𝘥𝘴—𝘥𝘦𝘴𝘱𝘪𝘵𝘦 𝘣𝘰𝘵𝘩
𝘣𝘦𝘪𝘯𝘨 𝘪𝘮𝘢𝘨𝘦-𝘩𝘦𝘢𝘷𝘺 𝘥𝘰𝘮𝘢𝘪𝘯𝘴?
Pathology has converged on large DINOv2-based models. EO has a mix of smaller models and the occasional larger model trained with diverse SSL objectives.
Let's unpack why 👇.
1️⃣ Built by Startups vs Labs
𝐏𝐚𝐭𝐡𝐨𝐥𝐨𝐠𝐲: Industry-led (PathAI, Owkin, Paige) — private, product-focused
𝐄𝐎: Often academic — open-source, exploratory
𝐒𝐨𝐦𝐞 𝐞𝐱𝐜𝐞𝐩𝐭𝐢𝐨𝐧𝐬: UNI and CONCH (Harvard) in pathology, AlphaEarth (Google) in EO
EO encourages experimentation. Pathology trends toward convergence.
2️⃣ Weak Supervision vs Scene
Understanding
𝐏𝐚𝐭𝐡𝐨𝐥𝐨𝐠𝐲: Weak slide-level labels, little fine-tuning
𝐄𝐎: Stronger labels, frequent fine-tuning
Pathology needs robust out-of-the-box representations. EO benefits from flexible backbones.
3️⃣ Proprietary vs Open Datasets
𝐏𝐚𝐭𝐡𝐨𝐥𝐨𝐠𝐲: Siloed and private
𝐄𝐎: Open, global-scale data (Sentinel and Landsat)
EO teams can iterate on both data and SSL. Pathology teams are more limited and less motivated to experiment.
4️⃣ DINOv2 vs Diverse Objectives
𝐏𝐚𝐭𝐡𝐨𝐥𝐨𝐠𝐲: DINOv2 dominates — generalizes well without fine-tuning
𝐄𝐎: Uses masked autoencoders, contrastive, distillation, hybrids
DINOv2 suits cases where fine-tuning isn't
feasible. EO models are frequently fine-tuned.
5️⃣ Product Focus vs Research Mindset
𝐄𝐎: Academic incentives = publication, exploration
𝐏𝐚𝐭𝐡𝐨𝐥𝐨𝐠𝐲: Startup incentives = speed, product fit
Research seeks understanding. Startups seek outcomes.
📌📌 𝐓𝐡𝐞𝐬𝐞 𝐜𝐨𝐧𝐭𝐫𝐚𝐬𝐭𝐬 𝐡𝐚𝐯𝐞𝐧'𝐭 𝐠𝐨𝐭𝐭𝐞𝐧 𝐦𝐮𝐜𝐡 𝐚𝐭𝐭𝐞𝐧𝐭𝐢𝐨𝐧—𝐛𝐮𝐭 𝐢𝐭 𝐞𝐱𝐩𝐥𝐚𝐢𝐧𝐬 𝐚 𝐥𝐨𝐭 𝐚𝐛𝐨𝐮𝐭 𝐡𝐨𝐰
𝐟𝐨𝐮𝐧𝐝𝐚𝐭𝐢𝐨𝐧 𝐦𝐨𝐝𝐞𝐥𝐬 𝐞𝐯𝐨𝐥𝐯𝐞.
So What's Driving The Gap?
• Use case structure
• Model design
• Data availability
• Fine-tuning feasibility
• Organizational incentives
✅ Different needs → different FM design choices.
🔍 These factors shape what 'good' looks like for architecture, objectives, and evaluation.
💬 Curious to hear your take:
• Seen similar patterns elsewhere?
• Are we heading for convergence—or more fragmentation?
|
|
|
|
|
|
 |
Research: EO Embeddings
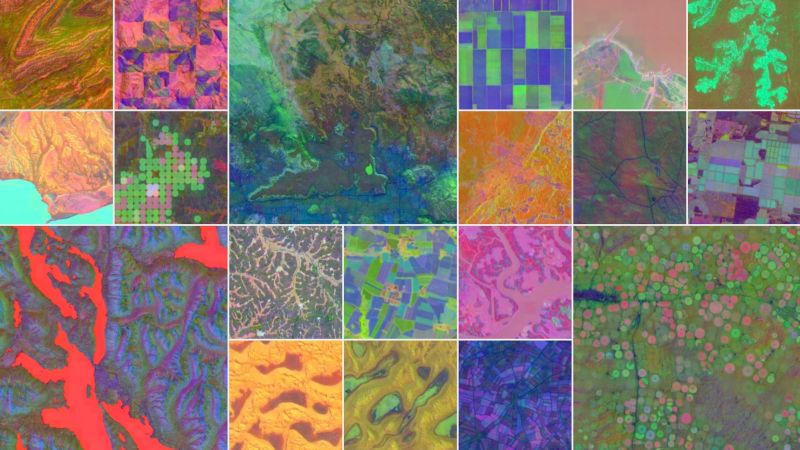
AlphaEarth Foundations: An embedding field model for accurate and efficient global mapping from sparse label data
𝘈 𝘯𝘦𝘸 𝘦𝘮𝘣𝘦𝘥𝘥𝘪𝘯𝘨-𝘧𝘰𝘤𝘶𝘴𝘦𝘥 𝘮𝘰𝘥𝘦𝘭 𝘥𝘦𝘴𝘪𝘨𝘯𝘦𝘥 𝘧𝘰𝘳 𝘨𝘭𝘰𝘣𝘢𝘭-𝘴𝘤𝘢𝘭𝘦 𝘦𝘢𝘳𝘵𝘩 𝘰𝘣𝘴𝘦𝘳𝘷𝘢𝘵𝘪𝘰𝘯—𝘸𝘪𝘵𝘩𝘰𝘶𝘵 𝘳𝘦𝘵𝘳𝘢𝘪𝘯𝘪𝘯𝘨.
Google just
released AlphaEarth—a geospatial foundation model trained on 3B+ observations across sensors, time, and terrain. And it doesn’t need fine-tuning. You simply work with the embeddings.
𝐀𝐥𝐩𝐡𝐚𝐄𝐚𝐫𝐭𝐡 𝐢𝐬𝐧’𝐭 𝐣𝐮𝐬𝐭 𝐚𝐧𝐨𝐭𝐡𝐞𝐫 𝐄𝐎 𝐟𝐨𝐮𝐧𝐝𝐚𝐭𝐢𝐨𝐧 𝐦𝐨𝐝𝐞𝐥—it’𝐬 𝐛𝐮𝐢𝐥𝐭 𝐟𝐨𝐫 𝐮𝐬𝐚𝐛𝐥𝐞, 𝐠𝐞𝐧𝐞𝐫𝐚𝐥𝐢𝐳𝐚𝐛𝐥𝐞 𝐞𝐦𝐛𝐞𝐝𝐝𝐢𝐧𝐠𝐬. Rather than optimizing for fine-tuning
performance, it generates 𝘤𝘰𝘮𝘱𝘢𝘤𝘵, 𝘩𝘪𝘨𝘩-𝘳𝘦𝘴𝘰𝘭𝘶𝘵𝘪𝘰𝘯 𝘦𝘮𝘣𝘦𝘥𝘥𝘪𝘯𝘨𝘴 that outperform handcrafted features and learned baselines in classification, regression, and change detection—even in low-label settings.
AlphaEarth was trained with a distillation objective—an approach known to produce more task-transferable features.
Most recent EO models (like Prithvi) are built around reconstruction objectives like masked autoencoding. These can perform well—but typically only after task-specific fine-tuning.
Prithvi struggled in frozen settings—sometimes even trailing an ImageNet-pretrained ViT with no EO knowledge.
That’s not surprising. It wasn’t designed to produce usable features
out-of-the-box.
AlphaEarth flips the script:
✅ 𝘊𝘰𝘯𝘵𝘪𝘯𝘶𝘰𝘶𝘴, 𝘴𝘱𝘢𝘵𝘪𝘢𝘭𝘭𝘺 𝘥𝘦𝘯𝘴𝘦 𝘦𝘮𝘣𝘦𝘥𝘥𝘪𝘯𝘨𝘴
✅ 𝘛𝘢𝘴𝘬-𝘢𝘨𝘯𝘰𝘴𝘵𝘪𝘤 𝘳𝘦𝘱𝘳𝘦𝘴𝘦𝘯𝘵𝘢𝘵𝘪𝘰𝘯𝘴
✅ 𝘋𝘦𝘴𝘪𝘨𝘯𝘦𝘥 𝘧𝘰𝘳 𝘪𝘯𝘧𝘦𝘳𝘦𝘯𝘤𝘦-𝘧𝘪𝘳𝘴𝘵 𝘸𝘰𝘳𝘬𝘧𝘭𝘰𝘸𝘴
For teams operationalizing EO
and remote sensing:
• Lowers the barrier to experimentation
• Robust to diverse inputs and sparse labels
• Enables prototyping without costly retraining
🔍 𝐖𝐡𝐚𝐭 𝐌𝐚𝐤𝐞𝐬 𝐀𝐄𝐅 𝐃𝐢𝐟𝐟𝐞𝐫𝐞𝐧𝐭?
• 🧠 𝐄𝐦𝐛𝐞𝐝𝐝𝐢𝐧𝐠-𝐟𝐢𝐫𝐬𝐭 – works out of the box
• 💾 𝐓𝐢𝐧𝐲 𝐛𝐮𝐭 𝐩𝐨𝐰𝐞𝐫𝐟𝐮𝐥 – ~64 bytes, 10 m resolution
• 🛰️ 𝐌𝐮𝐥𝐭𝐢𝐦𝐨𝐝𝐚𝐥 + 𝐭𝐞𝐦𝐩𝐨𝐫𝐚𝐥 – learns from sparse, diverse data
• 🌍
𝐓𝐫𝐚𝐢𝐧𝐞𝐝 𝐚𝐭 𝐬𝐜𝐚𝐥𝐞 – 3B+ observations; annual global layers
🧠 𝐖𝐡𝐞𝐧 𝐭𝐨 𝐔𝐬𝐞 𝐀𝐄𝐅
AEF won’t fit every scenario—fine-tuned models still shine in high-resource or fine-grained settings. But AEF is ideal when:
• Labels are sparse or expensive
• You can’t retrain for every task
• You need generalization across location and time
Example: mapping tree species or land-use change
👉 𝘊𝘢𝘯 𝘺𝘰𝘶 𝘴𝘰𝘭𝘷𝘦 𝘺𝘰𝘶𝘳 𝘵𝘢𝘴𝘬 𝘸𝘪𝘵𝘩 𝘧𝘳𝘰𝘻𝘦𝘯
𝘦𝘮𝘣𝘦𝘥𝘥𝘪𝘯𝘨𝘴 𝘭𝘪𝘬𝘦 𝘈𝘭𝘱𝘩𝘢𝘌𝘢𝘳𝘵𝘩? If yes, you’ve just skipped months of modeling—and moved closer to impact.
Blog
|
|
|
|
|
|
 |
Research: VLM for Pathology
Multimodal Whole Slide Foundation Model for Pathology
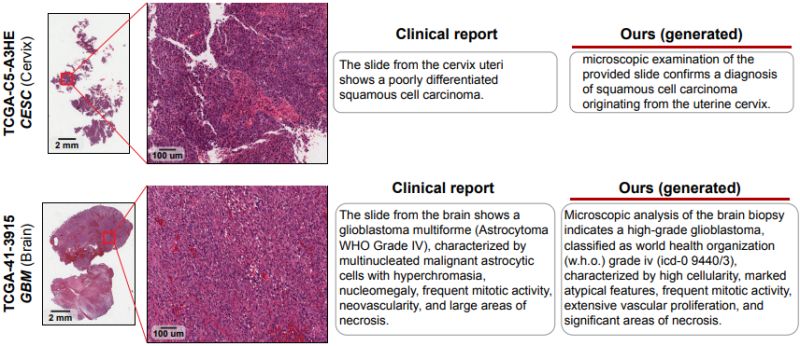
What if an AI could analyze entire histopathology slides and automatically generate detailed pathology reports that rival those written by experts? Tong Ding et al. just introduced TITAN, a multimodal foundation model that combines visual analysis of whole slide images with natural language understanding to transform computational pathology.
𝐓𝐡𝐞 𝐰𝐡𝐨𝐥𝐞 𝐬𝐥𝐢𝐝𝐞 𝐜𝐡𝐚𝐥𝐥𝐞𝐧𝐠𝐞: While existing pathology AI models excel at analyzing small tissue patches, they struggle to understand patterns across entire slides - the way pathologists actually work. Most foundation models are
trained on tiny regions and lack the context needed for comprehensive slide-level diagnosis, especially when integrating visual findings with the rich information found in pathology reports.
𝐊𝐞𝐲 𝐢𝐧𝐧𝐨𝐯𝐚𝐭𝐢𝐨𝐧𝐬:
• Scale: Trained on 335k whole slide images with 423k synthetic captions - the largest multimodal pathology dataset to date
• Novel 3-stage training: First self-supervised visual learning, then vision-language alignment with AI-generated captions, and finally alignment with pathology reports
• Zero-shot capability: Can analyze new cases without additional training or clinical labels
• Multi-resolution processing: Handles high-resolution whole slides (8,192 × 8,192 pixels) rather than just small patches
𝐖𝐡𝐲 𝐭𝐡𝐢𝐬
𝐦𝐚𝐭𝐭𝐞𝐫𝐬: TITAN can generate pathology reports, perform zero-shot classification, and enable cross-modal retrieval between histology slides and clinical reports. This could significantly improve diagnostic consistency, reduce pathologist workload, and provide crucial support in resource-limited settings where expert pathologists are scarce - particularly valuable for rare diseases where training data is typically insufficient.
The work represents a major step toward AI systems that can understand medical images with the nuanced reasoning typically required of human experts.
|
|
|
|
|
|
Enjoy this newsletter? Here are more things you might find helpful:
Team Workshop: Harnessing the Power of Foundation Models for Pathology - Ready to unlock new possibilities for your pathology AI product development? Join me for an exclusive 90 minute workshop designed to catapult your team’s model development.
Schedule now
|
|
|
Did someone forward this email to you, and you want to sign up for more? Subscribe to future emails
This email was sent to _t.e.s.t_@example.com. Want to change to a different address? Update subscription
Want to get off this list? Unsubscribe
My postal address: Pixel Scientia Labs, LLC, PO Box 98412, Raleigh, NC 27624, United States
|
|
|
|
|