 |
Insights: Translating AI in Pathology
The Illusion of Readiness
Your model might be brilliant in the lab—and blind in the clinic.
𝐖𝐡𝐲 𝐘𝐨𝐮𝐫 𝐏𝐚𝐭𝐜𝐡‑𝐋𝐞𝐯𝐞𝐥 𝐂𝐥𝐚𝐬𝐬𝐢𝐟𝐢𝐞𝐫 𝐈𝐬𝐧’𝐭 𝐑𝐞𝐚𝐝𝐲 𝐟𝐨𝐫 𝐭𝐡𝐞 𝐋𝐚𝐛
You’ve trained a model on patch-level histology images. It hits 95% accuracy. It looks great on paper.
But then… it flops when deployed in a real pathology lab.
Why?
Because patch-level performance doesn't guarantee whole-slide
reliability. Clinical slides bring a different world:
- Scanner variability
- Staining inconsistency
- Tissue and imaging artifacts
- Diagnostic ambiguity
- Contextual reasoning across regions
What worked on clean, cropped, color-normalized patches often falls apart when scaled to full slides—especially from a different lab or device.
Most real-world failures don’t come from bad models.
They come from models that were never tested under real-world conditions.
Take these two examples:
🔹 Scanner Shift Surprise: A breast cancer model trained on one scanner’s images showed high performance—but dropped by 30% when deployed with a different scanner brand, even though the images looked nearly identical to the human eye. It had learned subtle scanner-specific cues without developers realizing it.
🔹 Unseen Tissue Types: A colon cancer model performed well on resection specimens but failed when applied
to biopsy samples. The tissue structure and context were different—and the model had never seen biopsies during training.
𝐓𝐚𝐤𝐞𝐚𝐰𝐚𝐲: Without attention to generalizability, your model may never deliver clinical value—no matter how well it performs in validation.
📣 𝐘𝐨𝐮𝐫 𝐭𝐮𝐫𝐧: What’s the most surprising way your model failed outside the lab?
|
|
|
|
|
|
 |
Research: Agents for Pathology
PathFinder: A Multi-Modal Multi-Agent System for Medical Diagnostic Decision-Making Applied to Histopathology
Pathologists spend hours navigating gigapixel-scale tissue images, carefully examining suspicious regions and gathering evidence to make critical cancer diagnoses. It's a methodical, iterative process that requires enormous expertise—and it's becoming unsustainable as cancer cases rise globally.
New research by Ghezloo et al. introduces PathFinder, a multi-agent AI system that mimics this exact diagnostic workflow, achieving something remarkable: it's the first AI to outperform average pathologist performance in melanoma classification.
𝐖𝐡𝐲 𝐭𝐡𝐢𝐬 𝐦𝐚𝐭𝐭𝐞𝐫𝐬:
Traditional AI approaches analyze tissue patches independently,
missing the holistic reasoning that makes expert pathologists so effective. They look at pieces but lose the forest for the trees. Real diagnosis requires navigating the image strategically, building evidence, and synthesizing findings—exactly what PathFinder does.
𝐊𝐞𝐲 𝐢𝐧𝐧𝐨𝐯𝐚𝐭𝐢𝐨𝐧𝐬:
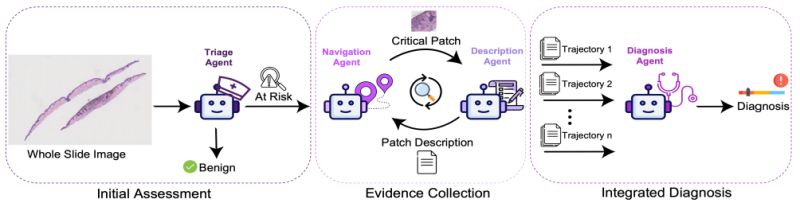
◦ 𝐅𝐨𝐮𝐫-𝐚𝐠𝐞𝐧𝐭 𝐚𝐫𝐜𝐡𝐢𝐭𝐞𝐜𝐭𝐮𝐫𝐞: Triage Agent classifies risk level, Navigation Agent identifies suspicious regions, Description Agent analyzes patches in natural language, and Diagnosis Agent synthesizes final conclusions
◦ 𝐈𝐭𝐞𝐫𝐚𝐭𝐢𝐯𝐞 𝐞𝐯𝐢𝐝𝐞𝐧𝐜𝐞
𝐠𝐚𝐭𝐡𝐞𝐫𝐢𝐧𝐠: Each agent's findings inform the next, creating a feedback loop that refines focus with each examination cycle
◦ 𝐈𝐧𝐭𝐞𝐫𝐩𝐫𝐞𝐭𝐚𝐛𝐥𝐞 𝐫𝐞𝐚𝐬𝐨𝐧𝐢𝐧𝐠: Natural language descriptions of diagnostically relevant patches provide clear explanations for decisions
𝐓𝐡𝐞 𝐫𝐞𝐬𝐮𝐥𝐭𝐬:
PathFinder achieved 74% accuracy on the challenging M-Path melanoma dataset—a 9% improvement over the 65% average human pathologist performance and 8% better than previous state-of-the-art methods. Expert pathologists rated the system's patch descriptions as comparable to GPT-4o quality.
𝐖𝐡𝐲 𝐭𝐡𝐢𝐬
𝐦𝐚𝐭𝐭𝐞𝐫𝐬:
This isn't just about automation—it's about augmenting human expertise in areas where diagnostic accuracy literally saves lives. The multi-agent approach offers something traditional AI lacks: explainable reasoning that pathologists can review, validate, and learn from.
As cancer cases continue rising globally, systems like PathFinder could help democratize access to expert-level diagnostic capabilities while maintaining the interpretability that medical professionals demand.
|
|
|
|
|
|
 |
Research: Foundation Models for Agriculture
Few-Shot Adaptation of Grounding DINO for Agricultural Domain
Advanced vision-language models like Grounding DINO excel at detecting objects using text prompts in natural images. "Grounding" refers to the AI's ability to connect language descriptions with specific visual regions - essentially linking words to pixels. But try asking them to identify individual wheat heads in a crowded field or distinguish crop leaves from weeds - they often fail dramatically.
Rajhans Singh et al. tackle this challenge by developing a few-shot adaptation method that makes these powerful models work effectively for agricultural applications.
𝗧𝗵𝗲 𝗣𝗿𝗼𝗺𝗽𝘁 𝗣𝗿𝗼𝗯𝗹𝗲𝗺 𝗶𝗻
𝗔𝗴𝗿𝗶𝗰𝘂𝗹𝘁𝘂𝗿𝗲:
Grounding DINO's zero-shot approach relies on text prompts, but agricultural scenarios present unique challenges:
• 𝗢𝗰𝗰𝗹𝘂𝗱𝗲𝗱 𝗮𝗻𝗱 𝗰𝗹𝘂𝘁𝘁𝗲𝗿𝗲𝗱 𝗼𝗯𝗷𝗲𝗰𝘁𝘀: Individual leaves in dense plants or wheat heads partially hidden by foliage are hard to describe precisely in text
• 𝗩𝗶𝘀𝘂𝗮𝗹𝗹𝘆 𝘀𝗶𝗺𝗶𝗹𝗮𝗿 𝗰𝗹𝗮𝘀𝘀𝗲𝘀: Young crop plants and certain weeds look nearly identical - difficult to distinguish through text alone
• 𝗖𝗼𝗺𝗽𝗹𝗲𝘅
𝗰𝗮𝘁𝗲𝗴𝗼𝗿𝗶𝗲𝘀: How do you write a prompt for "other insects" that captures hundreds of species while excluding specific ones?
𝗧𝗵𝗲 𝗙𝗲𝘄-𝗦𝗵𝗼𝘁 𝗦𝗼𝗹𝘂𝘁𝗶𝗼𝗻:
Instead of struggling with text prompts, the researchers developed an elegant adaptation:
• 𝗥𝗲𝗺𝗼𝘃𝗲 𝘁𝗲𝘅𝘁 𝗰𝗼𝗺𝗽𝗹𝗲𝘅𝗶𝘁𝘆: Eliminate the BERT text encoder entirely from Grounding DINO
• 𝗥𝗮𝗻𝗱𝗼𝗺 𝗲𝗺𝗯𝗲𝗱𝗱𝗶𝗻𝗴
𝗶𝗻𝗶𝘁𝗶𝗮𝗹𝗶𝘇𝗮𝘁𝗶𝗼𝗻: Replace text features with randomly initialized trainable embeddings that match BERT's output dimensions
• 𝗠𝗶𝗻𝗶𝗺𝗮𝗹 𝘁𝗿𝗮𝗶𝗻𝗶𝗻𝗴: Learn these embeddings using just a few labeled images and hundreds of training iterations
𝗜𝗺𝗽𝗿𝗲𝘀𝘀𝗶𝘃𝗲 𝗥𝗲𝘀𝘂𝗹𝘁𝘀:
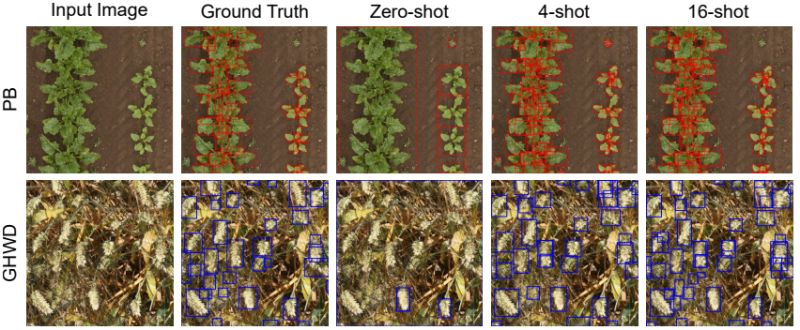
The approach was tested across eight diverse agricultural datasets:
• ~24% higher performance than fully fine-tuned YOLO models when training data is limited
• ~10% improvement over state-of-the-art methods in remote sensing tasks
• Significant gains from just 4-16 training images across crop-weed detection, fruit counting, and
pest identification
• Works for both object detection and instance segmentation when combined with SAM2
𝗪𝗵𝘆 𝗧𝗵𝗶𝘀 𝗠𝗮𝘁𝘁𝗲𝗿𝘀:
Manual annotation of agricultural datasets is expensive and time-intensive. This approach could:
• Automate annotation pipelines for agricultural AI development
• Enable rapid deployment of object detection for new crops or pests
• Reduce the barrier to entry for precision agriculture technologies
• Support farmers and researchers with limited labeled data
This work makes powerful vision-language models practically useful for agriculture - where visual complexity often exceeds what language can easily describe.
|
|
|
|
|
|
Enjoy this newsletter? Here are more things you might find helpful:
Team Workshop: Harnessing the Power of Foundation Models for Pathology - Ready to unlock new possibilities for your pathology AI product development? Join me for an exclusive 90 minute workshop designed to catapult your team’s model development.
Schedule now
|
|
|
Did someone forward this email to you, and you want to sign up for more? Subscribe to future emails
This email was sent to _t.e.s.t_@example.com. Want to change to a different address? Update subscription
Want to get off this list? Unsubscribe
My postal address: Pixel Scientia Labs, LLC, PO Box 98412, Raleigh, NC 27624, United States
|
|
|
|